As an embryo develops, it needs to produce a myriad of cell types, precisely organized in space and in exact proportions. This extraordinary feat requires cells to interpret complex histories of signals from a small number of conserved pathways, such as BMP, Wnt, and Nodal. These signaling histories profoundly influence cell fate decisions, yet capturing these histories remains challenging, particularly in mammalian systems where live imaging is limited by size and developmental timescales. To overcome this challenge, we have developed innovative molecular recording methods that encode signaling events directly into cellular genomes, enabling reconstruction of cell signaling histories at single-cell resolution, long after these events have occurred.
Our lab is now expanding these molecular recording technologies, enhancing their sensitivity and dynamic range to robustly capture the diverse intensities and temporal sequences of signals encountered during early embryogenesis, stem cell differentiation, and tissue formation. Coupling these approaches with spatial transcriptomics allows us to correlate signaling histories with cell fate and tissue organization in situ. Through interdisciplinary integration of gene editing, quantitative imaging, epigenomic analyses, computational modeling, and synthetic biology, we aim to uncover the fundamental principles governing how cells coordinate their developmental decisions over time. Ultimately, our goal is to leverage these insights to inform novel strategies for tissue engineering and to open new therapeutic avenues for diseases driven by dysregulated developmental signaling.
Research Directions
Question: How signals that cells receive early on relate to their eventual fate?
Tool: Molecular recording
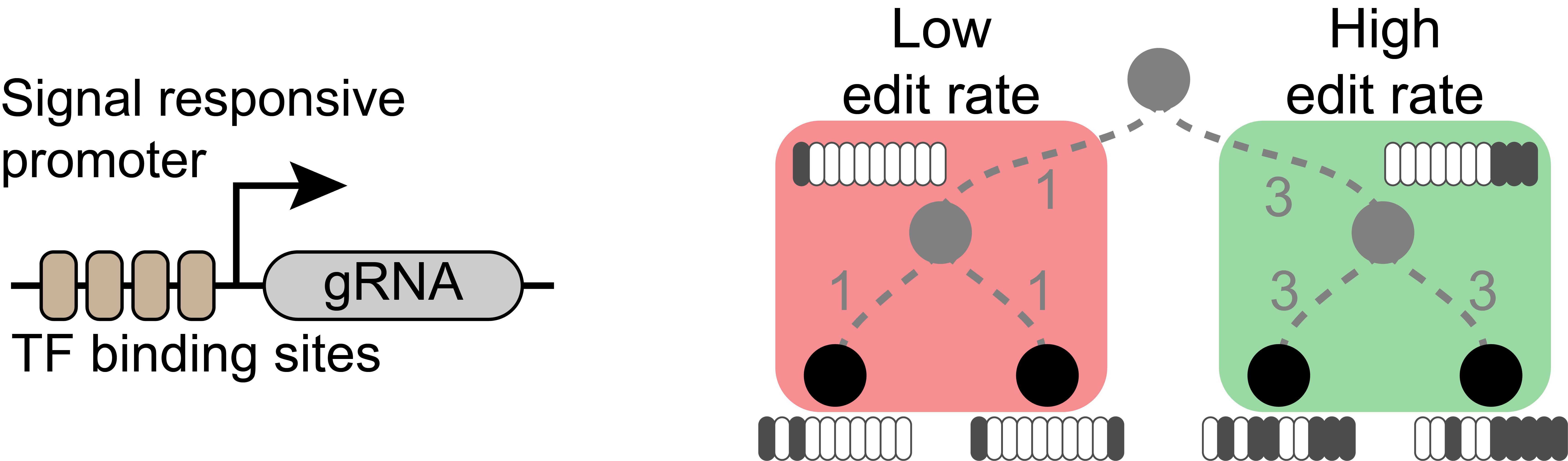
Cells use signaling pathways to orchestrate their behavior during development. A surprisingly limited number of key signaling pathways are used to elicit numerous cellular responses. Cells use intensity and order of these signals to interpret their meaning and make appropriate decisions.
We use genome engineering and imaging based barcoding to link the intensity and order of signals that a cell receives to the phenotype of its progeny. If mutations in an array of barcodes are made at a rate proportional to the intensity of a signal, a permanent record of that signal will be made in the genome of the cells. Mutation rate in our genetic recording system depends on the expression level of the CRISPR base editor and the guide RNA. By coupling the expression of base editor or gRNA to the signal of interest we can develop a molecular recording system and explore how early events in the progenitors are linked to cellular diversification. The same scheme can also be used to study how aberrant signaling leads to cancer and developmental disorders.
Question: To what extent the lineage of a cell affect its fate?
Tool: Lineage recording
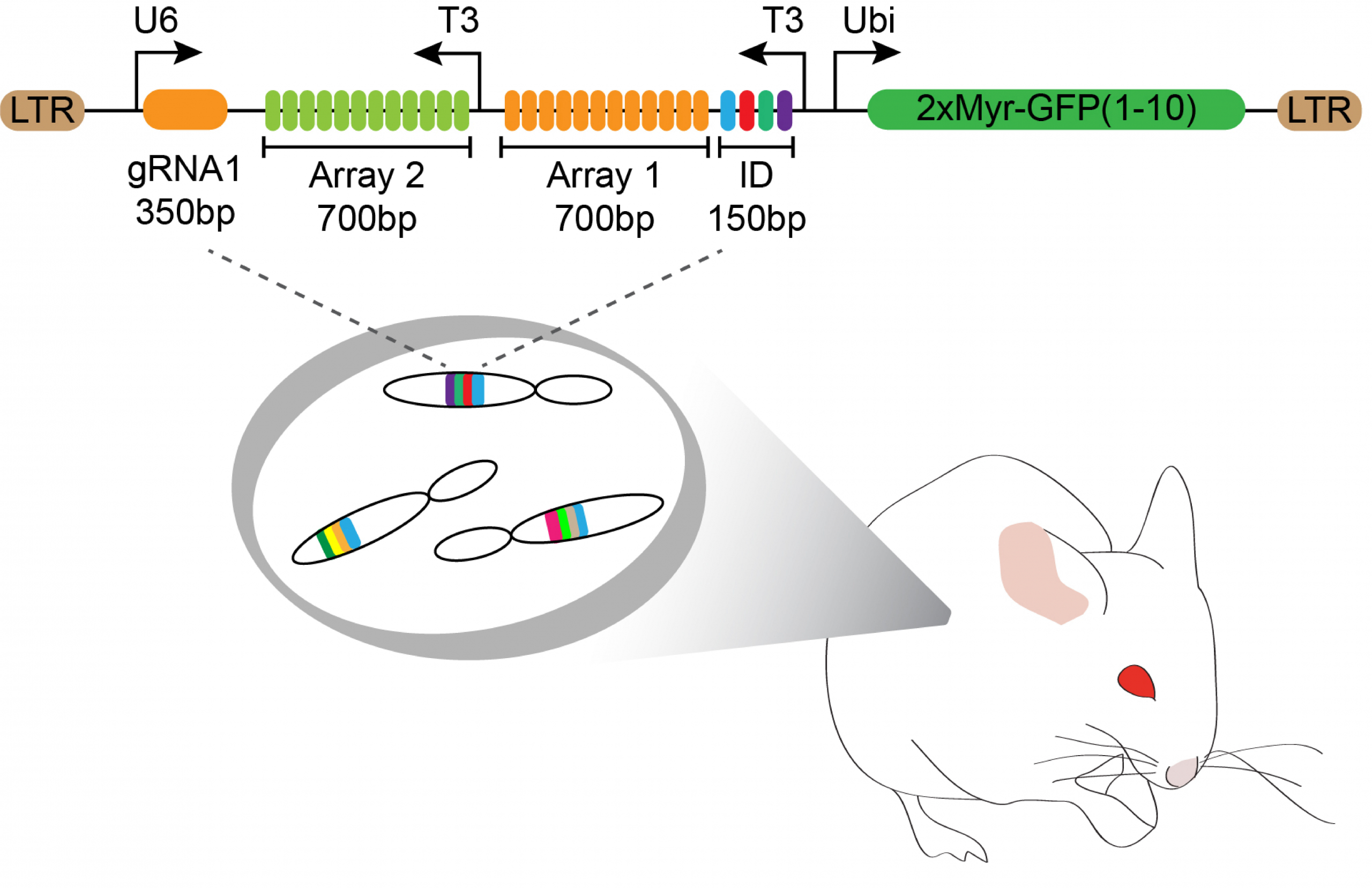
Neuroblasts in Drosophila and C. elegans produce neurons and glia in a stereotypical sequence instructed by cell fate regulators. Neurons and glia of the mouse retina are also born in an evolutionarily conserved order, although with significant overlap between the cell types. It is tempting to think that mammalian retinal progenitors also transition through a reproducible sequence of competence states and the overlap is only caused by asynchrony between lineages. However, this process in vertebrates may be regulated differently and allow for more stochastic cell fate choices.
Knowing the set of cell types that each progenitor makes is not enough to resolve this type of issues. We need to know the sequence of cell fate decisions along individual lineage trees. In systems where time lapse microscopy is not feasible, either due to inaccessibility of the tissue or the time scale of development, reconstruction of lineage trees poses a significant challenge. Lineage recording, in which mutations accumulate in heritable barcodes as cells grow, offers a promising solution to this problem. We have developed a synthetic system in which a CRISPR base editor can gradually make stochastic mutations in an array of compact image readable barcodes. This approach allows reconstruction of lineage relationships between cells. Analysis of the reconstructed trees will reveal the extent to which programmed lineages versus stochastic processes operate in cell fate decisions.